
Stuart Tompson, PhD and colleagues at the University of Wisconsin—Madison are exploring a novel cell profiling approach to understand Primary Congenital Glaucoma (PCG). PCG is a severe pediatric eye disease affecting 1 in 10,000 infants in genetically diverse populations. Patients with PCG may have a cloudy cornea – the front of the eye that is normally clear – and enlarged eyes. It results from elevated eye pressure that damages the optic nerve and can potentially lead to blindness. Current treatment involves surgery, and, in some cases, eye removal.
The disease is not, at present, fully understood. Tompson, a researcher in the Department of Ophthalmology and Visual Sciences, is hoping to change that.
Research to date has shown that the disease is caused by a failure to correctly develop the structures of the aqueous humor outflow (AHO) pathway, which are crucial for regulating drainage of fluid from the eye. Mutations in four genes (CYP1B1, LTBP2, TEK, and ANGPT1) are currently known to cause the disease, and account for 25% of cases.
With funding from the NIH/NEI, Tompson is utilizing a new “cell” sequencing method to shed light on which genes are important for the development of AHO tissues – and identify the underlying gene mutations in the remaining 75% of cases.
“We have hypothesized that the elusive genes underlying PCG are also critical for and therefore expressed during the development of the AHO pathway,” Tompson said. “However, the scientific community currently lacks a comprehensive knowledge of the genes that are expressed during the formation of these important ocular structures.”
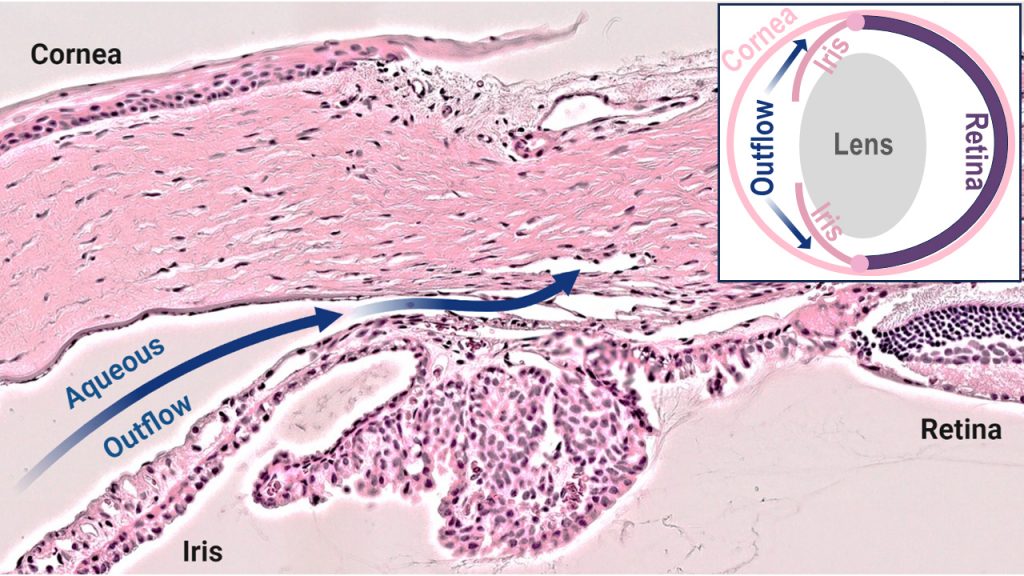
Until recently, profiling the developing AHO pathway was not feasible because the diminutive and complex structures contain a diverse array of cell types. Dr. Tompson is now leveraging single-cell RNA sequencing (scRNAseq) technology to broadly profile the gene expression in each of these individual cell types as the tissues form. His research utilizes rat tissues, which mirror human AHO pathway development.
“We will generate datasets from male and female tissues at various developmental timepoints,” Tompson said. “This will enable us to identify differentially expressed genes and pathways specific to each cell type, developmental stage, and gender. We will then use this resource to prioritize genes as candidates for disease-causing variants within existing exome sequencing data from molecularly unsolved PCG cases.”
Ultimately, these studies aim to unveil novel molecular mechanisms underlying PCG, paving the way for gene-based diagnostic testing, improved genetic counseling, enhanced clinical management, and the development of more effective drug treatments focused on disease biology rather than surgical interventions.